Virtual companions, real responsibility
Large Language Models in Medicine:
The digital transformation of medicine depends on reliable processing and linking of medical workflows. Large Language Models (LLMs) contribute significantly to better structuring, categorization and interpretation of medical information.However, these models suffer from weaknesses such as the generation of plausible but incorrect information, known as hallucination, and the ability to produce different outputs for the same input, i.e. non-determinism. In a paper published in the Nature Portfolio journal “npj Digital Medicine”, researchers from the EKFZ for Digital Health at TU Dresden and the University of Chile discuss possible solutions. The combination of LLMs and knowledge graphs (KGs), a form of “Retrieval Augmented Generation”, could make the models more reliable, robust and reproducible.
The reliable recording of medical information and exchange between different systems (interoperability) remains a significant challenge in healthcare and is commonly referred to as medicine’s ‘communication problem’. Approaches to address this issue are medical ontologies and knowledge graphs (KGs). Medical ontologies function like dictionaries for medical terms, which help to categorize and define medical concepts. However, due to the fact that in human language terms can have different meanings depending on the context, these ontologies are often ambiguous. The word “cold” for instance can refer to body temperature, environmental conditions or the clinical syndrome “acute rhinitis”. KGs are organized networks, that connect different medical concepts and their relationships. For instance, the term “COVID-19” in a KG, could be connected to “fever” through a link labeled “has symptom”. KGs facilitate understanding and processing of medical information, but face similar challenges as medical ontologies.
To address these shortcomings, the researchers from Dresden and Santiago de Chile propose combining LLMs with KGs, leveraging their respective strengths. This combination is a form of “Retrieval Augmented Generation”. It provides structured reasoning and could help reduce model bias and deliver more reliable, accurate, and reproducible results. These approaches would be more compatible with regulatory approval pathways than LLMs alone.
“The combination of large language models and knowledge graphs is a way of linking existing medical knowledge with the cognitive capabilities of large language models. We are only at the beginning of a very exciting development here,” says Prof. Jakob N. Kather, Professor of Clinical Artificial Intelligence at TU Dresden and oncologist at Dresden University Hospital Carl Gustav Carus.
The authors discuss different approaches to combining LLMs with KGs. They suggest that this could also facilitate the development of robust digital twins of patients, in the form of individual structured health records that enable personalized diagnosis.
“Although regulatory challenges remain, healthcare professionals graduating today can anticipate access to compatible and advanced clinical information summarization tools that were previously unimaginable just five years ago. In addition, approaches combining large language models with knowledge graphs are more likely to achieve early approval in conservative regulatory pathways”, says Prof. Stephen Gilbert, Professor of Medical Device Regulatory Science at TU Dresden.
More News
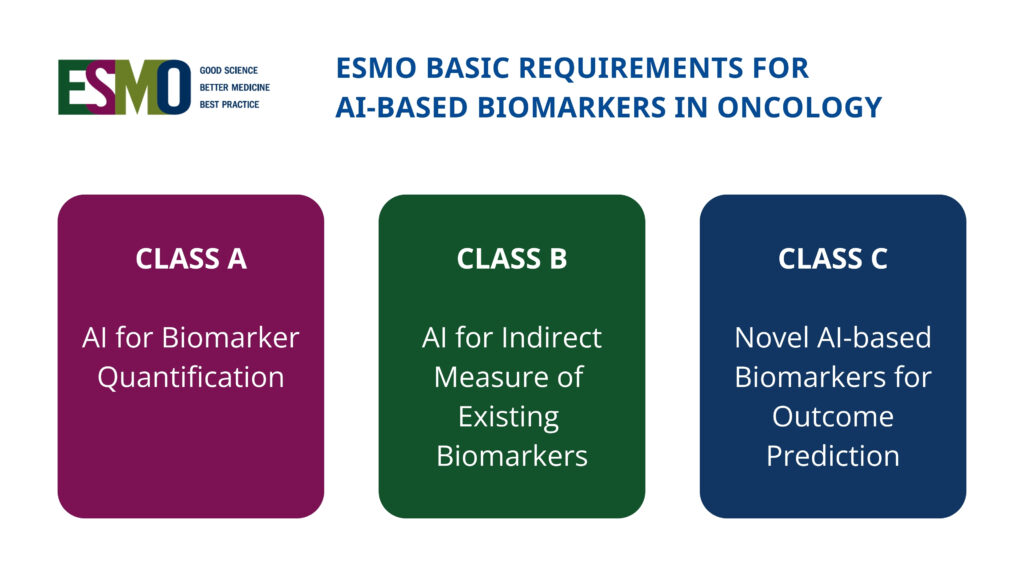
New international framework defines standards for AI-based biomarkers in oncology (EBAI)
New ESMO Guidelines: Safe use of large language models in oncology